How to Read H-nmr and C-nmr to Detrmine Structure
NMR - Estimation
- Page ID
- 1812
Nuclear Magnetic Resonance (NMR) interpretation plays a pivotal role in molecular identifications. As interpreting NMR spectra, the structure of an unknown compound, as well as known structures, can be assigned by several factors such as chemic shift, spin multiplicity, coupling constants, and integration. This Module focuses on the near of import oneH and xiiiC NMR spectra to detect out structure even though at that place are diverse kinds of NMR spectra such as 14N, 19F, and 31P. NMR spectrum shows that ten- axis is chemical shift in ppm. It as well contains integral areas, splitting pattern, and coupling constant.
Strategy for Solving Structure
Here is the general strategy for solving structure with NMR:
- Molecular formula is determined by chemical analysis such as elementary analysis
- Double-bond equivalent (also known as Caste of Unsaturation) is calculated by a simple equation to estimate the number of the multiple bonds and rings. Information technology assumes that oxygen (O) and sulfur (Due south) are ignored and halogen (Cl, Br) and nitrogen is replaced past CH. The resulting empirical formula is CaHb
.jpg?revision=1)
- Structure fragmentation is adamant by chemical shift, spin multiplicity, integral (peak surface area), and coupling constants (\(^1J\), \(^2J\))
- Molecular skeleton is built upwards using 2-dimensional NMR spectroscopy.
- Relative configuration is predicted past coupling constant (3J).
1H NMR
Chemical Shift
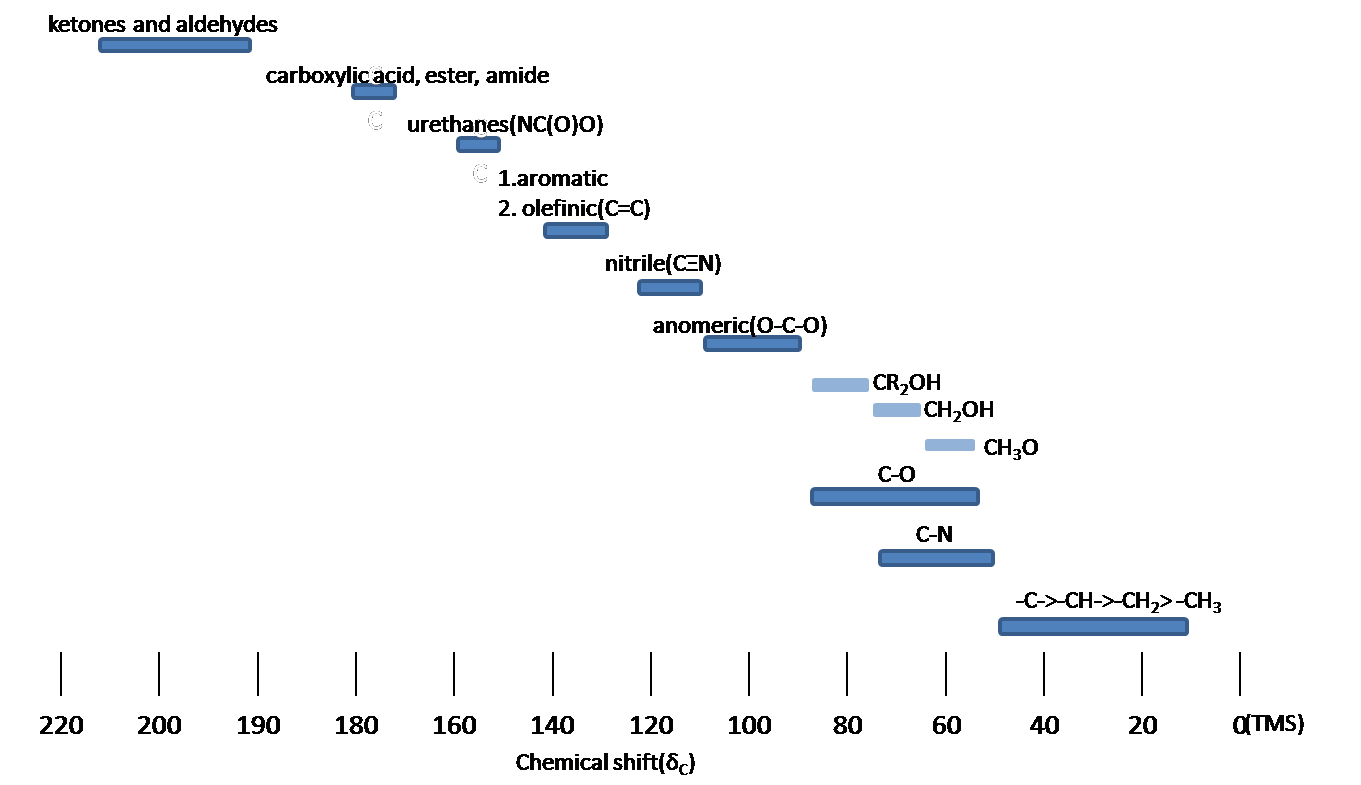
Chemical shift is associated with the Larmor frequency of a nuclear spin to its chemic surround. Tetramethylsilane (TMS, \(\ce{(CH3)4Si}\)) is generally used equally an internal standard to determine chemical shift of compounds: δTMS=0 ppm. In other words, frequencies for chemicals are measured for a 1H or 13C nucleus of a sample from the aneH or 13C resonance of TMS. It is important to understand tendency of chemical shift in terms of NMR interpretation. The proton NMR chemical shift is affect by nearness to electronegative atoms (O, N, element of group vii.) and unsaturated groups (C=C,C=O, aromatic). Electronegative groups motion to the down field (left; increment in ppm). Unsaturated groups shift to downfield (left) when affecting nucleus is in the plane of the unsaturation, merely opposite shift takes place in the regions higher up and below this plane. 1H chemical shift play a part in identifying many functional groups. Figure \(\PageIndex{1}\). indicates important case to figure out the functional groups.
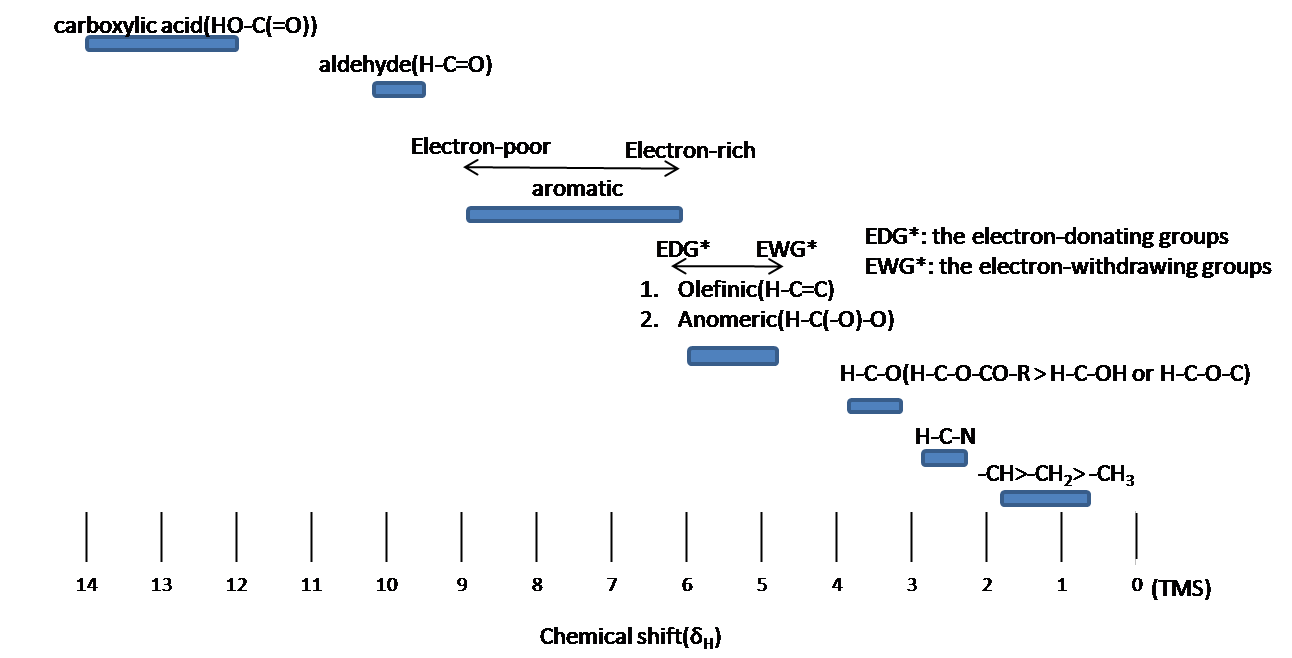
Chemical equivalence
Protons with Chemical equivalence has the same chemic shift due to symmetry within molecule (\(CH_3COCH_3\)) or fast rotation effectually single bond (-CH3; methyl groups).
Spin-Spin Splitting
Spin-Spin splitting means that an arresting top is split past more than than one "neighbor" proton. Splitting signals are separated to J Hz, where is chosen the coupling constant. The spitting is a very essential part to obtain exact information about the number of the neighboring protons. The maximum of distance for splitting is three bonds. Chemical equivalent protons practice non issue in spin-spin splitting. When a proton splits, the proton'due south chemical shift is determined in the center of the splitting lines.
Spin Multiplicity (Splitting pattern)
Spin Multiplicity plays a office in determining the number of neighboring protons. Here is a multiplicity rules: In instance of \(A_mB_n\) system, the multiplicity rule is that Nuclei of \(B\) element produce a splitting the \(A\) signal into \(nB+i\) lines. The general formula which applies to all nuclei is \(2_nI+1\), where \(I\) is the spin quantum number of the coupled element. The relative intensities of the each lines are given by the coefficients of the Pascal'due south triangle (Effigy \(\PageIndex{2}\)).
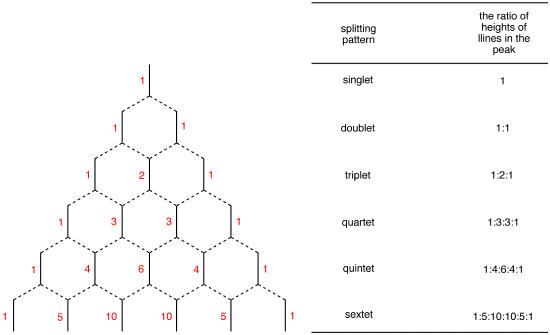
Beginning-society splitting design
The chemical shift deviation in Hertz between coupled protons in Hertz is much larger than the \(J\) coupling abiding:
\[ \dfrac{\Delta \nu }{J} \ge 8\]
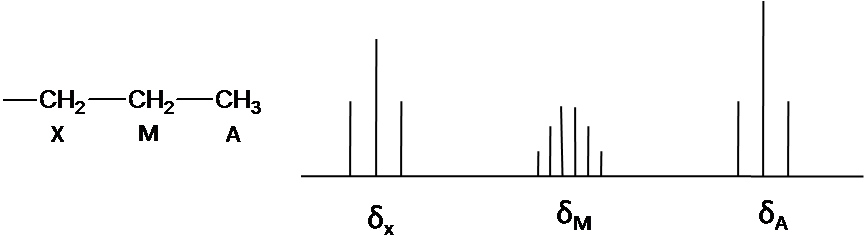
Where \(\Delta \nu\) is the difference of chemical shift. In other give-and-take, the proton is only coupled to other protons that are far away in chemical shift. The spectrum is called first-social club spectrum. The splitting pattern depends on the magnetic field. The second-gild splitting at the lower field can exist resolved into first-order splitting pattern at the high field. The first-guild splitting blueprint is allowed to multiplicity dominion (N+ane) and Pascal's triangle to determine splitting pattern and intensity distribution.
Instance \(\PageIndex{1}\)
The note is that structure organisation is A3Mtwo102. Ha and Hten has the triplet blueprint by Hm because of N+i rule. The signal of Hm is split into vi peaks past Hx and Ha (Figure3) The First guild pattern easily is predicted due to separation with equal splitting blueprint.
High-order splitting pattern
Loftier-order splitting pattern takes place when chemical shift deviation in Hertz is much less or the same that club of magnitude as the j coupling.
\[\frac{\Delta five}{J} \leq 10\]
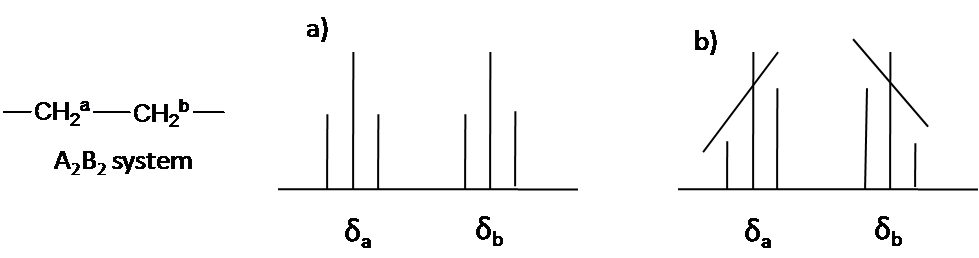
The 2nd order pattern is observed equally leaning of a classical blueprint: the inner peaks are taller and the outer peaks are shorter in case of AB system (Figure \(\PageIndex{4}\)). This is called the roof effect.
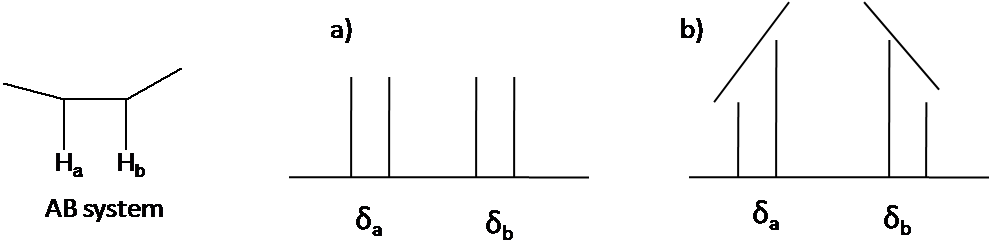
Hither is other system every bit an example: AtwoB2 (Figure \(\PageIndex{5}\)). The two triplet incline toward each other. Outer lines of the triplet are less than 1 in relative area and the inner lines are more than 1. The centre lines take relative area 2.
Coupling constant (J Value)
Coupling constant is the strength of the spin-spin splitting interaction and the distance betwixt the split lines. The value of distance is equal or different depending on the coupled nuclei. The coupling constants reflect the bonding environments of the coupled nuclei. Coupling constant is classified by the number of bonds:
Geminal proton-proton coupling (2JHH)
Germinal coupling generates through two bonds (Figure \(\PageIndex{six}\)). 2 proton having geminal coupling are non chemically equivalent. This coupling ranges from -20 to xl Hz. 2JHHdepends on hybridization of carbon atom and the bond angle and the substituent such every bit electronegative atoms. When Due south-character is increased, Geminal coupling constant is increased: 2Jsp1>2Jsp2>2Jsp3 The bond angle(HCH) gives rise to change 2JHH value and depend on the strain of the ring in the cyclic systems. Geminal coupling constant determines ring size. When bail angle is decreased, ring size is decreased so that geminal coupling abiding is more positive. If a atom is replace to an electronegative cantlet, Geminal coupling constant motion to positive value.

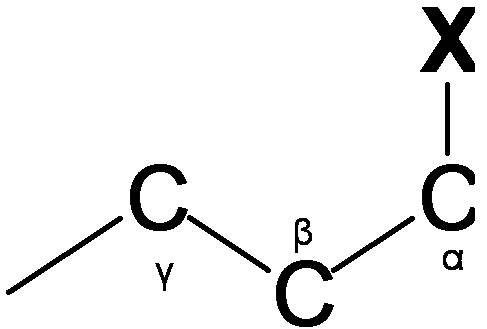
Vicinal proton-proton coupling (3JHH)
Vicinal coupling occurs though three bonds (Figure \(\PageIndex{7}\).). The Vicinal coupling is the nigh useful data of dihedral angle, leading to stereochemistry and conformation of molecules. Vicinal coupling constant always has the positive value and is affected by the dihedral angle (?;HCCH), the valence angle (?; HCC), the bond length of carbon-carbon, and the effects of electronegative atoms. Vicinal coupling constant depending on the dihedral angle (Effigy \(\PageIndex{viii}\)) is given by the Karplus equation.
\[^3 J=7.0-0.5 \cos \phi+4.five \cos ^{ii} \phi\]
When ? is the xco, vicinal coupling constant is zero. In improver, vicinal coupling abiding ranges from 8 to 10 Hz at the and ?=180o, where ?=0o and ?=180o ways that the coupled protons have cis and trans configuration, respectively.

The valence bending(?;Figure \(\PageIndex{viii}\)) also causes change of 3JHH value. Valence angle is related with band size. Typically, when the valence angle decreases, the coupling constant reduces. The distance betwixt the carbons atoms gives influences to vicinal coupling constant

The coupling constant increases with the subtract of bond length. Electronegative atoms touch on vicinal coupling constants and so that electronegative atoms subtract the vicinal coupling constants.
Integral
Integral is referred to integrated superlative area of 1H signals. The intensity is straight proportionally to the number of hydrogen.
xiiiC NMR
Chemic Shift
Spin-Spin splitting
Comparison the iH NMR, at that place is a large difference matter in the xiiiC NMR. The 13C- xiii C spin-spin splitting rarely exit between adjacent carbons considering 13C is naturally lower arable (1.ane%)
- xiiiC-1H Spin coupling: thirteenC-aneH Spin coupling provides useful information about the number of protons attached a carbon cantlet. In example of ane bond coupling (aneJCH), -CH, -CH2, and CHthree have respectively doublet, triplet, quartets for the 13C resonances in the spectrum. However, 13C-aneH Spin coupling has an disadvantage for 13C spectrum interpretation. 13C-aneH Spin coupling is hard to analyze and reveal structure due to a forest of overlapping peaks that result from 100% abundance of iH.
- Decoupling: Decoupling is the process of removing xiiiC-iH coupling interaction to simplify a spectrum and identify which pair of nuclei is involved in the J coupling. The decoupling 13C spectra shows simply 1 peak(singlet) for each unique carbon in the molecule(Figure \(\PageIndex{10}\).). Decoupling is performed past irradiating at the frequency of one proton with continuous low-ability RF.

- Distortionless enhancement by polarization transfer (DEPT): DEPT is used for distinguishing between a CH3 group, a CH2 group, and a CH group. The proton pulse is set at 45o, 90o, or 135o in the 3 divide experiments. The different pulses depend on the number of protons fastened to a carbon atom. Effigy \(\PageIndex{11}\). is an example near DEPT spectrum.

ii-dimensional NMR spectroscopy (COSY)
COSY stands for COrrelation SpectroscopY. COSY spectrum is more useful data nearly what is beingness correlated.
oneH-1H COSY (COrrelation SpectroscopY)
1H-1H COSY is used for clearly bespeak correlation with coupled protons. A point of entry into a COSY spectrum is one of the keys to predict data from information technology successfully. Relation of Coupling protons is adamant past cross peaks(correlation peaks) and in the COSY spectrum. In other words, Diagonal peaks by lines ar e coupled to each other. Effigy \(\PageIndex{12}\) indicates that at that place are correlation peaks betwixt proton H1 and Htwo as well as between Hii and H4. This means the H2 coupled to H1 and H4.

1H-13C COSY (HETCOR)
aneH-13C COSY is the heteronuclear correlation spectroscopy. The HETCOR spectrum is correlated 13C nuclei with directly attached protons. 1H-13C coupling is one bail. The cross peaks mean correlation between a proton and a carbon (Figure \(\PageIndex{xiii}\)). If a line does not have cross pinnacle, this means that this carbon atoms has no attached proton (e.g. a fourth carbon cantlet)

References
- Balc*, K., Basic p1 sH- and p13 sC-NMR spectroscopy. 1st ed.; Elsevier: Amsterdam ; Boston, 2005; p xii, 427.
- Breitmaier, Due east., Structure elucidation past NMR in organic chemistry : a applied guide. 3rd rev. ed.; Wiley: Chichester, West Sussex, England, 2002; p xii, 258.
- Jacobsen, N. East., NMR spectroscopy explained : simplified theory, applications and examples for organic chemistry and structural biology. Wiley-Interscience: Hoboken, Due north.J., 2007; p fifteen, 668.
- Silverstein, R. M.; Webster, F. X., Spectrometric identification of organic compounds. 6th ed.; Wiley: New York, 1998; p xiv, 482.
Exterior Links
- NMRShiftDB: a Free web database for NMR information : nmrshiftdb.chemie.uni-mainz.de/nmrshiftdb
- NMR database from ACD/LAbs : world wide web.acdlabs.com/products/spec_lab/exp_spectra/spec_libraries/aldrich.html
- NMR database from John Crerar Library : http://crerar.typepad.com/crerar_lib...h_ir_nmr_.html
Problems
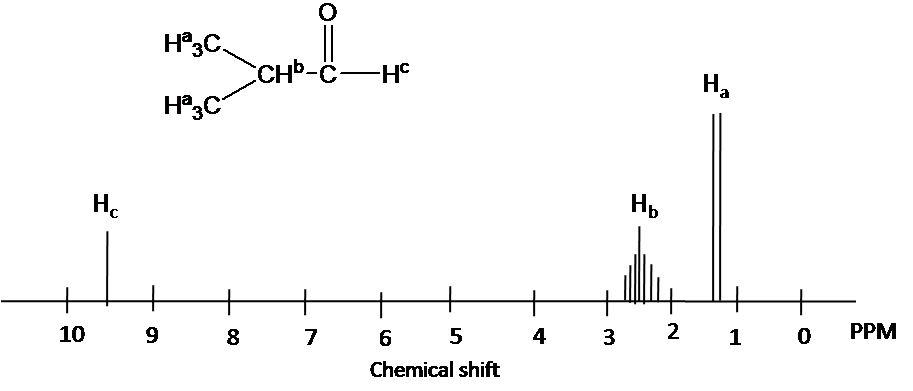
Describe the 1H NMR spectrum for 2-Hydroxypropane in CDCl3. Assume sufficient resolution to provide a first-order spectrum and ignore vicinal proton-proton coupling(3JHH)
Solution
1) the construction of 2-hydoroxyporpane is drawn

Effigy out which protons are chemically equivalent, i.e., ii methyl (-CHiii) groups are chemical equivalent.
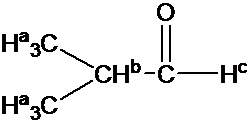
4) Splitting design is adamant by (N+one) rule: Ha is split up into two peaks past Hb(#of proton=1). Hb has the septet design by Ha (#of proton=6). Hc has i peak.(Note that Hc has doublet design by Hb due to vicinal proton-proton coupling.)
Contributors and Attributions
- You Jin Seo
Source: https://chem.libretexts.org/Bookshelves/Physical_and_Theoretical_Chemistry_Textbook_Maps/Supplemental_Modules_(Physical_and_Theoretical_Chemistry)/Spectroscopy/Magnetic_Resonance_Spectroscopies/Nuclear_Magnetic_Resonance/NMR%3A_Experimental/NMR_-_Interpretation
0 Response to "How to Read H-nmr and C-nmr to Detrmine Structure"
Post a Comment